ODISEI
Organoid-based drug efficacy and toxicity evaluation solution
ODISEI ONC
Oncology
ODISEI PET
ODISEI VIR
ODISEI NEP
ODISEIGUT
ODISEI CNS
ODISEI SKIN
ODISEI HEP
ODISEI Spatial
Overview

Advantages of Organoid Model
rate Mass
production Ethical
issue
Strategies: Basic Research
- Connecting genotypes to specific phenotypes
- Cancer initiation and progression
- Clonal evolution
- Cancer stem cell characterization
- Mechanism of drug response and resistance
Utilization Strategy for Each Stage of New Drug Development
Target Section
Lead Finding,
Prioritization Optimization
3. Drug efficacy Evaluate and optimize
Phase I
Phase II
Phase III
Organoid Establishment and Banking
- Skin & Hair
- Cerebral
- Midbrain
- Lacrimal Gland
- Salivary Gland
- Adenoid
- Tonsil
- Lung
- Heart
- Liver
- Stomach
- Kidney
- Intestine

- Prostate Cancer
- Hepatocellular Carcinoma
- Breast Cancer
- Pancreatic Cancer
- Renal Cell Carcinoma
- Endometrial Cancer
- Head & Neck Cancer
- Non-Small Cell Lung Cancer
- Cholangiocarcinoma
- Gastric Cancer
- Colorectal Cancer
- Ovarian Cancer
Available pre-clinical research
- Donor-matched immune microenvironment
- Various patient-derived organoid lineups
- High-resolution data
(HCS, FACS, NGS, scRNAseq, multiflex IHC) - Fully customized service
Available clinical data
- Basic clinical data
- Drug sensitivity prediction
- Patient follow-up data
- Clinical bioinformatics
(NGS, RNAseq, Spatial biology)
Applications
Client
Therapeutic Approaches
- Antibodies
- Anti-Cancer Drugs
- Vaccines
- Small Molecules
- Peptides
- Nucleosides
- Microbiome
- Virus
- Cosmetic Compounds
- Proinflammatory-Cytokines
- Proinflammatory-Chemokines
Disease Modeling
- Oncology Virus
- Oncology Skin
- Oncology Gut
- Cytotoxic T Cells
- Macrophages
- DCs
- Tregs
- NKs
- CAFs
- Skin
- Intestinal
- Liver
- Kidney
- Cardiac
- Lung
- Stomach
- Brain
Read-outs
- Morphology
- Cell viability
- Cell type composition
- Gene expression
- Mutation profiling
- Immune cell profiling
- Pathway analysis
- Cell to cell interaction
- Metabolism
Analysis
- High-content Screening
- Cell Viability
- Fluorescence-sorting
- Fluorescence-activated Cell Sorting
- Confocal Microscopy
- Immunohistochemistry
- Immunofluorescence
- Phenocycler-fusion
- (Multiplex-IHC)
- -CODEX, OPAL
- Bulk-RNAseq
- Single Cell RNAseq
- Whole Genome Sequencing
- MS/MS
Report
ODISEI ONC
High-Fidelity Preclinical Models
These models preserve the original tumor’s genetic and phenotypic characteristics, providing a more clinically relevant system than traditional 2D cell lines or animal models.
Extensive Organoid Biobanking
This resource enables the identification of drug candidates across diverse tumor profiles and informs strategic decision-making throughout the R&D pipeline.
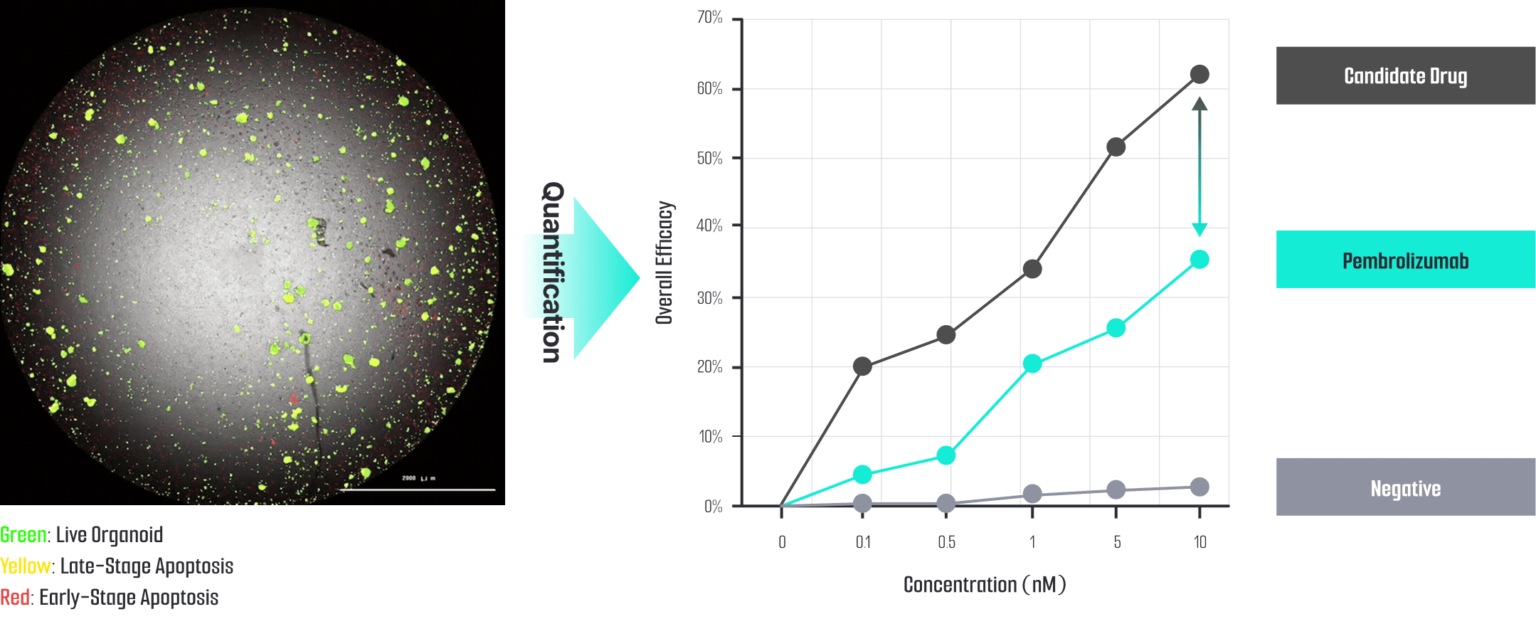
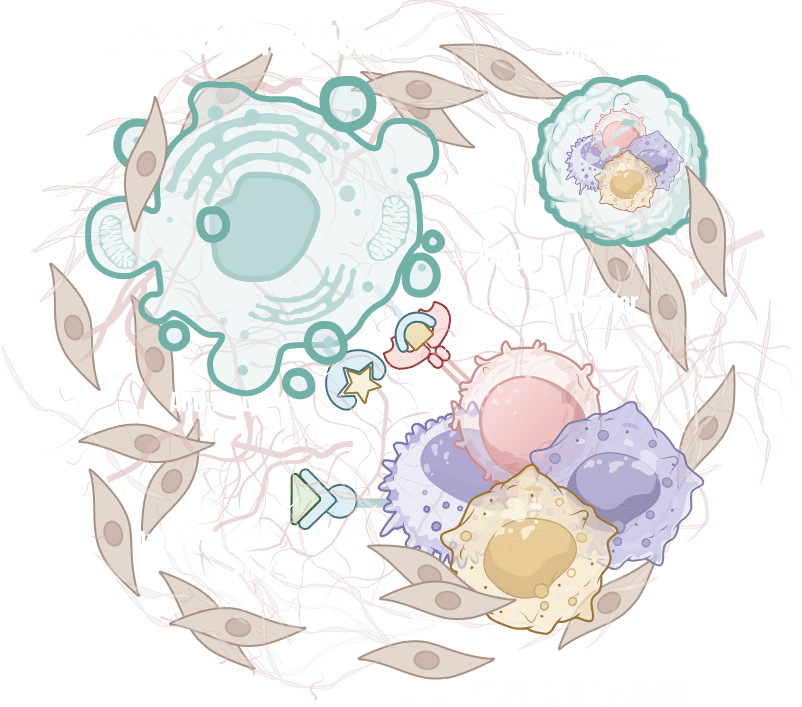
ODISEI-ONC : Immuno-Oncology Drug Evaluation Platform Using Tumor Organoids
Features
Immune-Microenvironment with Cytoxic T cells
Tumor-Infiltrating Lymphocyte (TIL)
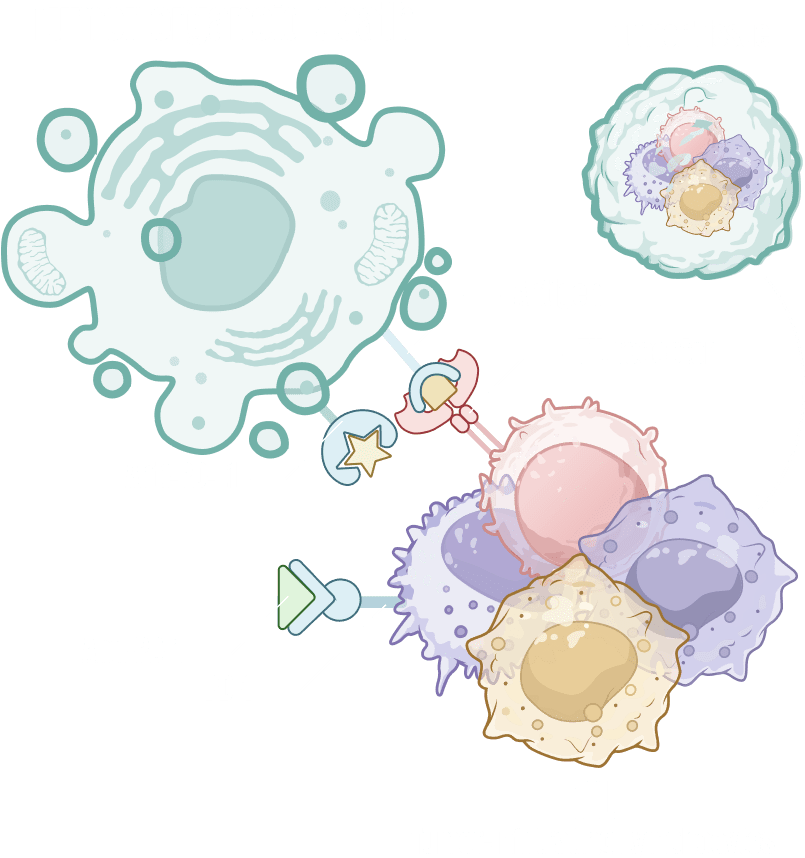
Regulatory T cell (Treg)
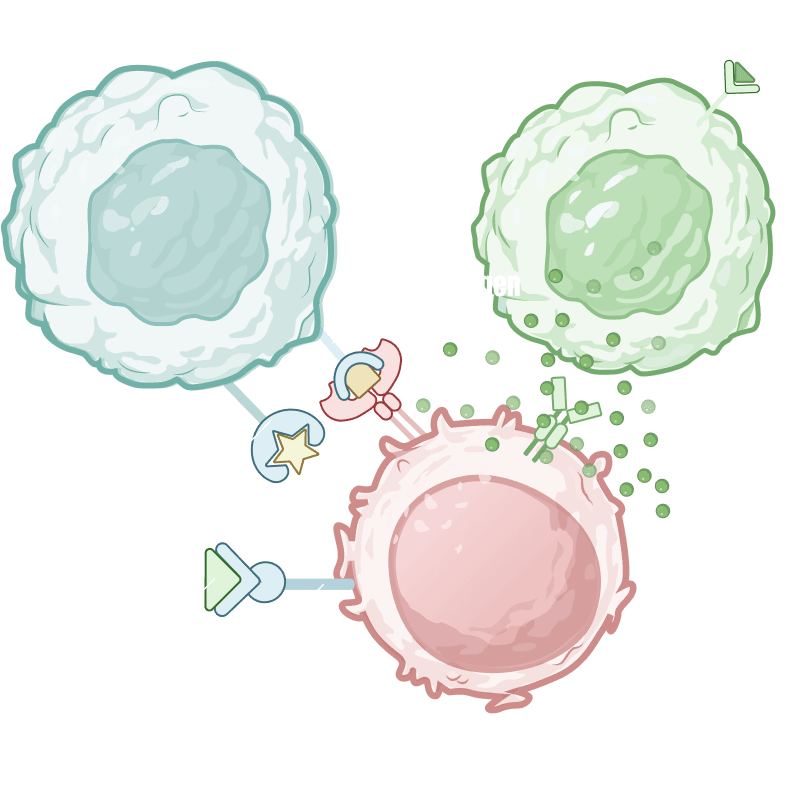
Macrophage
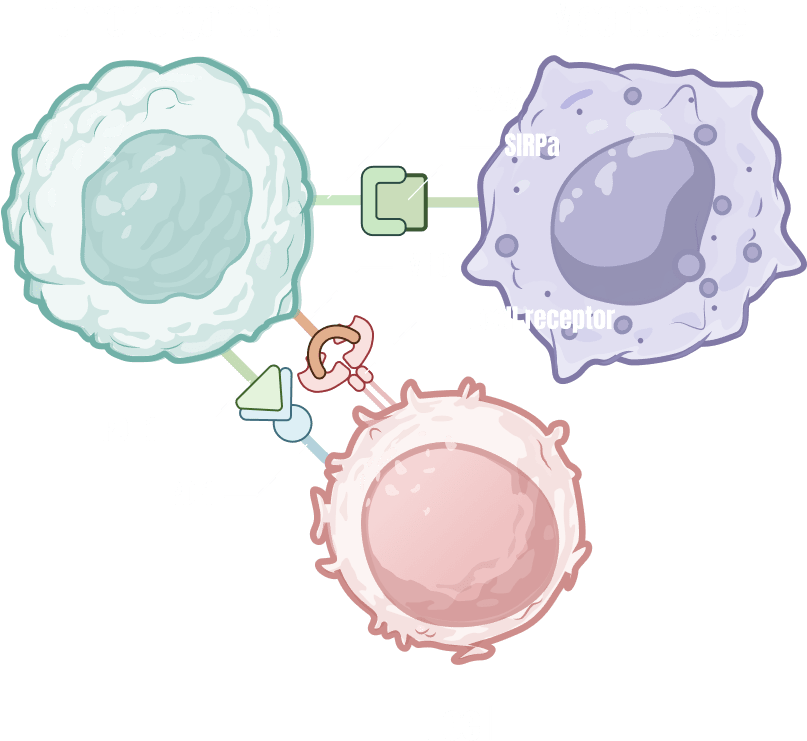
Cancer Associate Fibroblast (CAF)
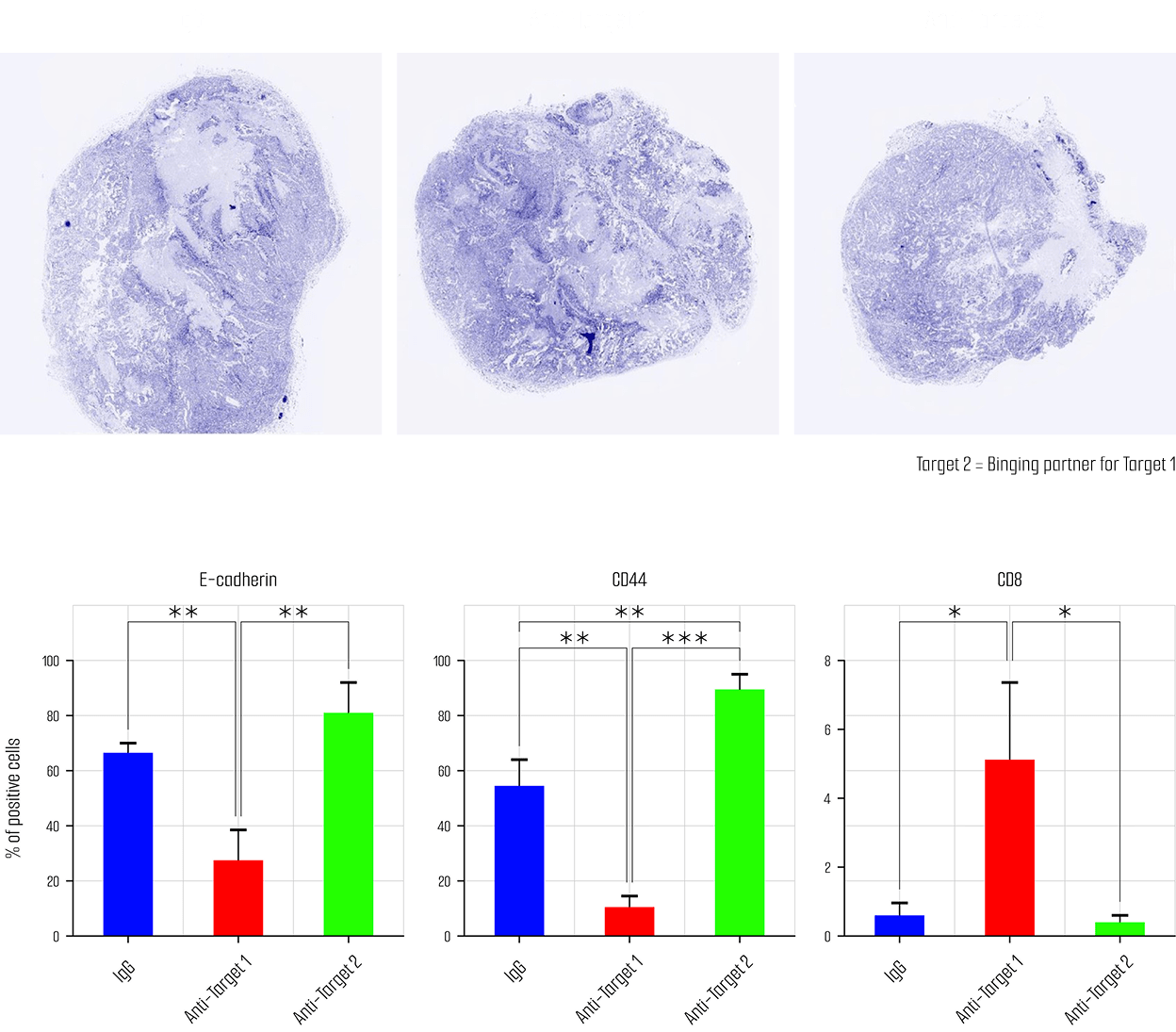
Validation Complete for caner organoid
- Clinical information
- Drug sensitivity test
- Genetic mutation (WES analysis)
- Specific cancer marker expression
(Immunohistochemistry, Multiflex)
WES analysis in pancreatic cancer organoid
Drug sensitivity test in pancreatic cancer organoid
ODISEI GUT
Generation of human intestinal organoid (hIO)
iPSC-derived intestinal organoids
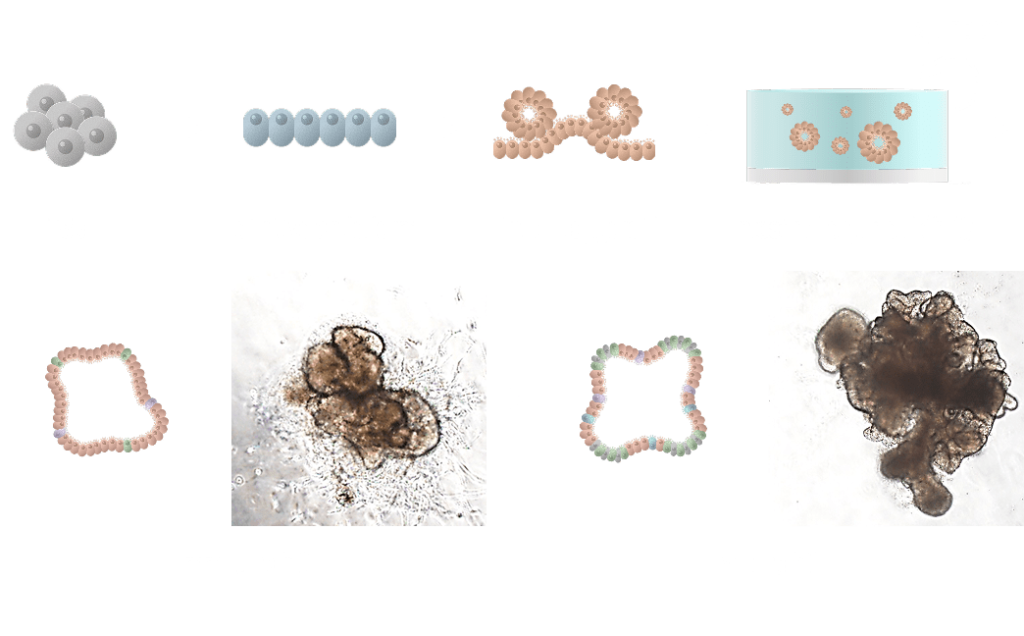
Tissue-derived intestinal organoids
Cellular and structural similarity
The small intestine is a crucial organ responsible for nutrient absorption, consisting of four layers: the mucosa, submucosa, muscularis propria, and serosa. Inside the small intestine, villi maximize nutrient absorption, while crypts serve as regions where intestinal epithelial cells proliferate. Additionally, microvilli further expand the surface area for absorption.
The major cell types include enterocytes responsible for nutrient uptake, goblet cells that secrete mucus, Paneth cells that produce antimicrobial peptides, enteroendocrine cells that release hormones, and intestinal stem cells that regenerate the intestinal epithelium.
Intestinal organoids have a structure and cellular composition similar to human intestinal tissue and mimic the functions of the small intestine.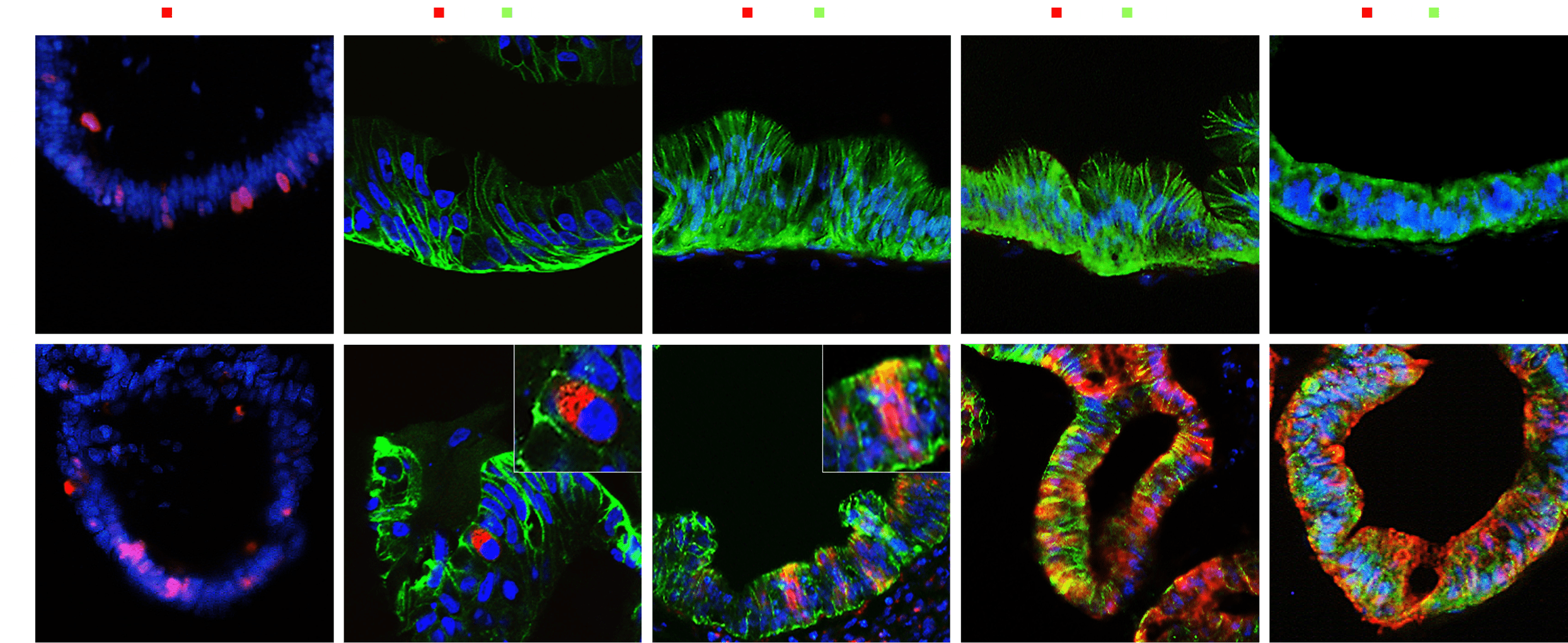
In our intestinal organoids, key markers for function and cell status have been identified, including Ki67, associated with cell proliferation; E-cadherin (Ecad), which maintains cell adhesion; OLFM4, a marker for intestinal stem cells; MUC13, involved in mucus secretion; and KRT20, indicative of mature intestinal epithelial cell differentiation.
Ki67 – Strongly expressed in proliferating cells within the crypts, indicating active cell proliferation
E-cadherin – Essential for maintaining adhesion between epithelial cells, ensuring structural integrity
OLFM4 – A key marker of intestinal stem cells, specifically expressed in the crypt region
MUC13 – Expressed in goblet cells and some enterocytes, contributing to the protection of the intestinal
KRT20 – Expressed in mature enterocytes, serving as an indicator of epithelial cell differentiation
Intestinal Disease & Function Models
LGS (Leaky Gut Syndrome) Model
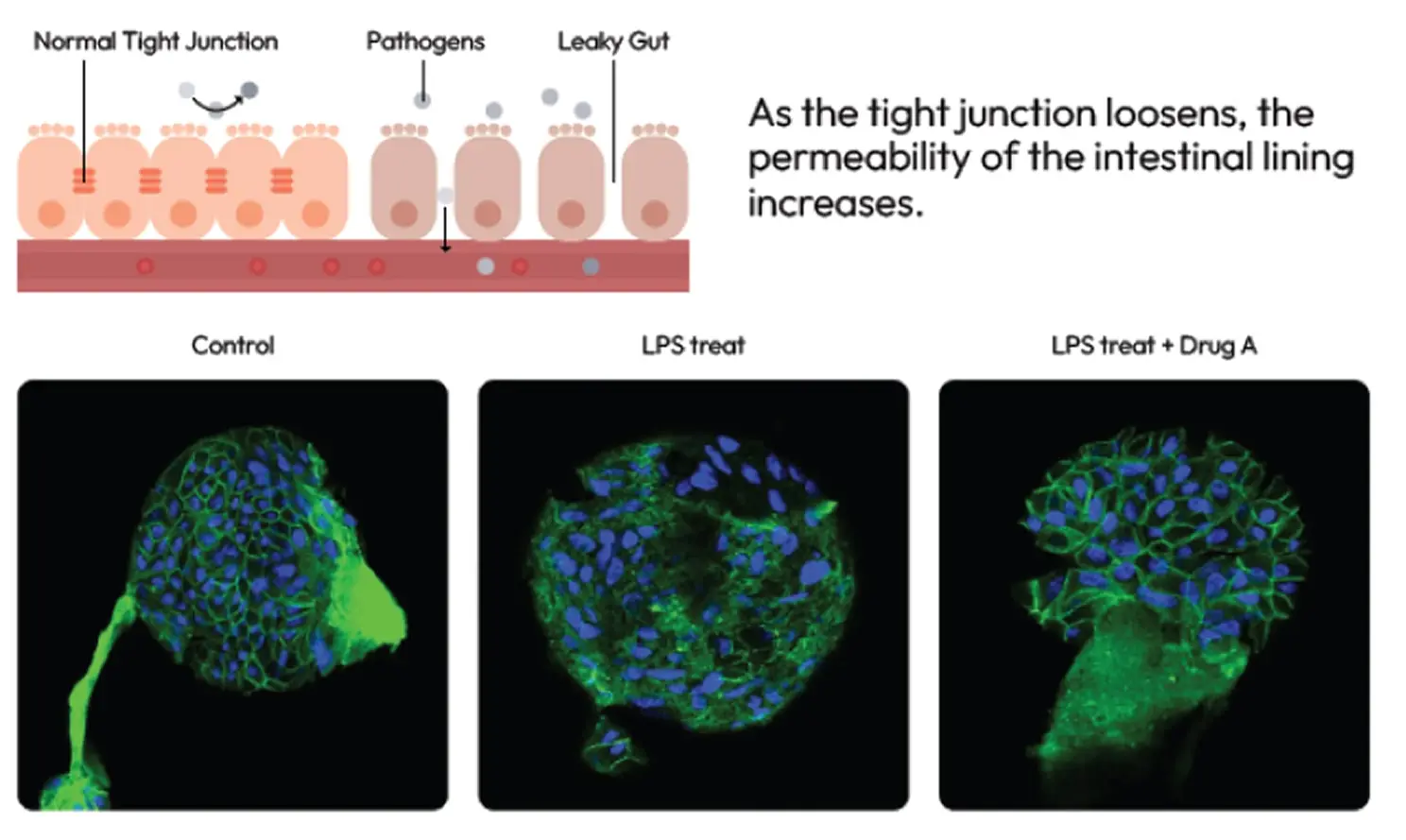
IBD (Inflammatory Bowel Disease) Model
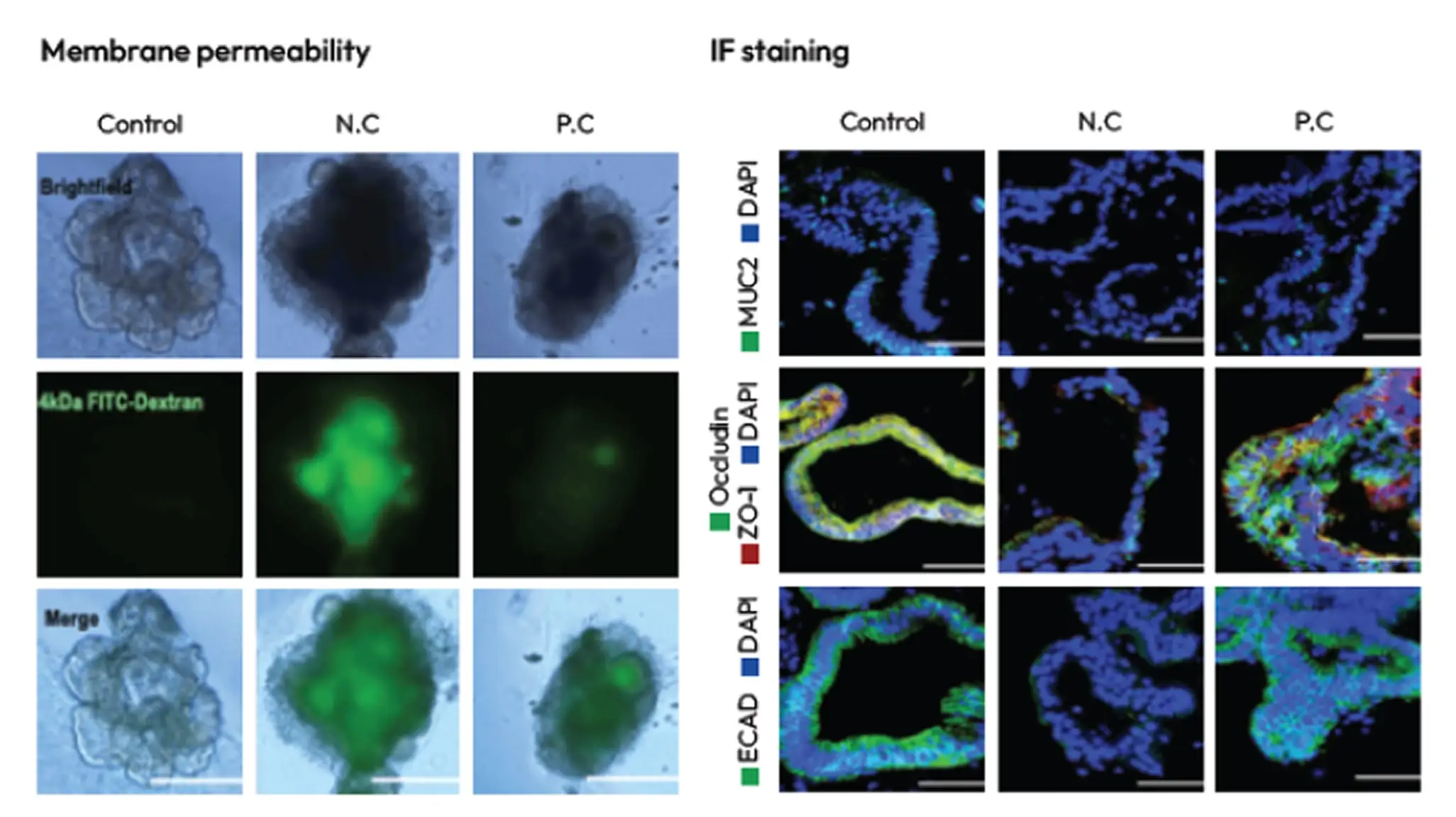
Drug Absorption and Permeability Evaluation Method
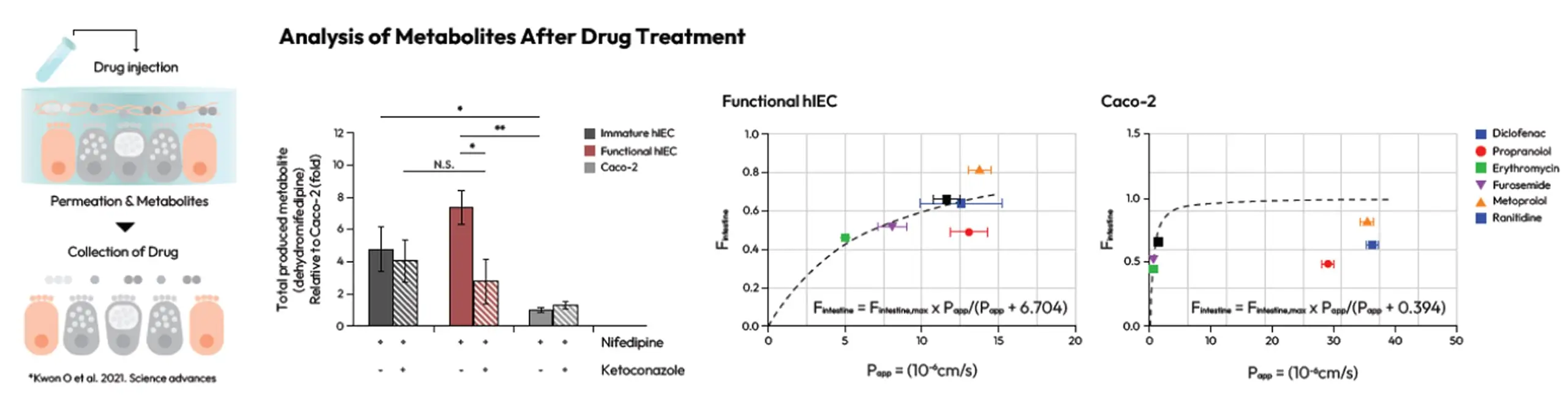
Analysis of Metabolites After Drug Treatment
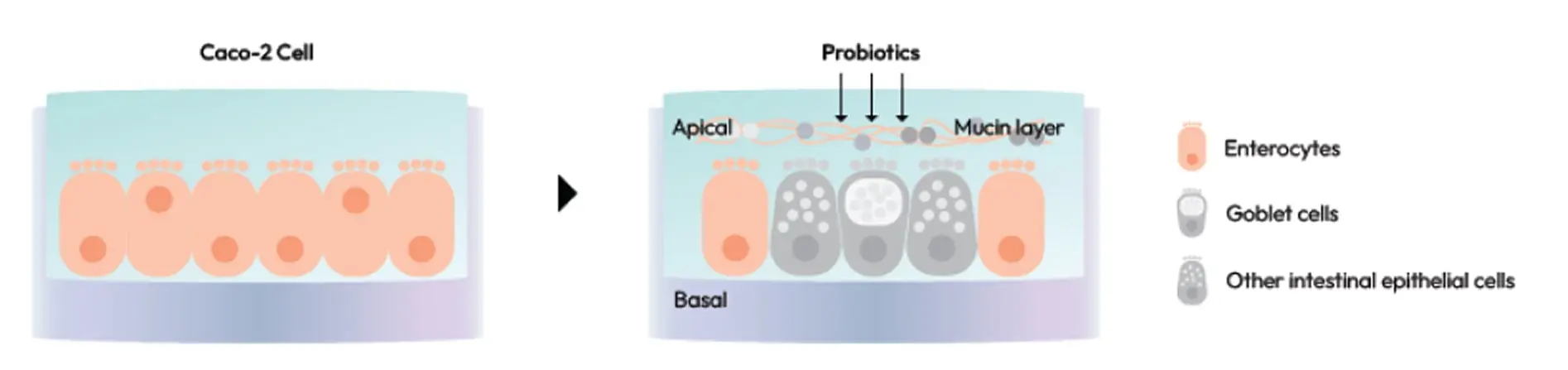
Microbiome & Probiotics
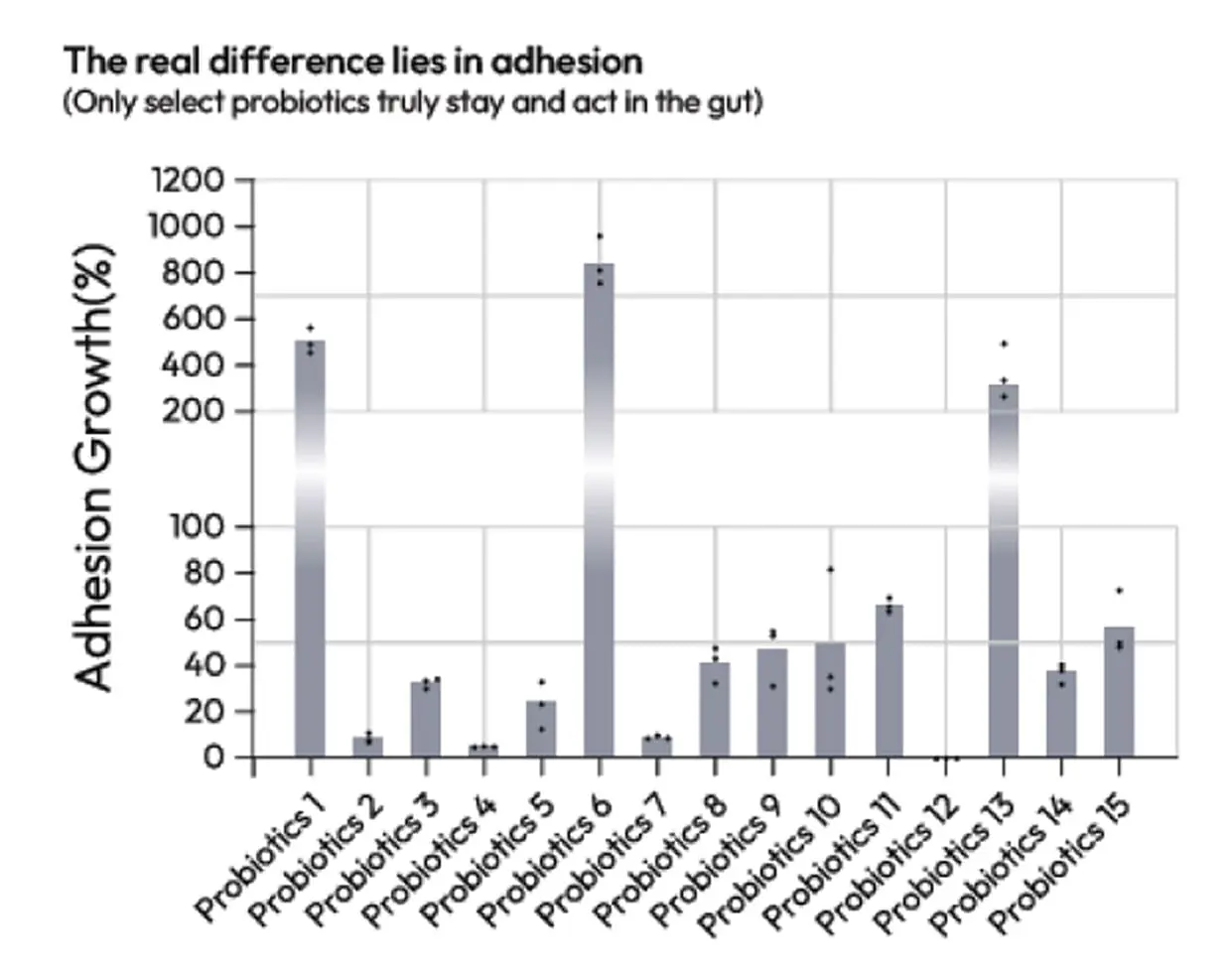
(Only select probiotics truly stay and act in the gut)
ODISEI SKIN
Generation of hair bearing skin organoid
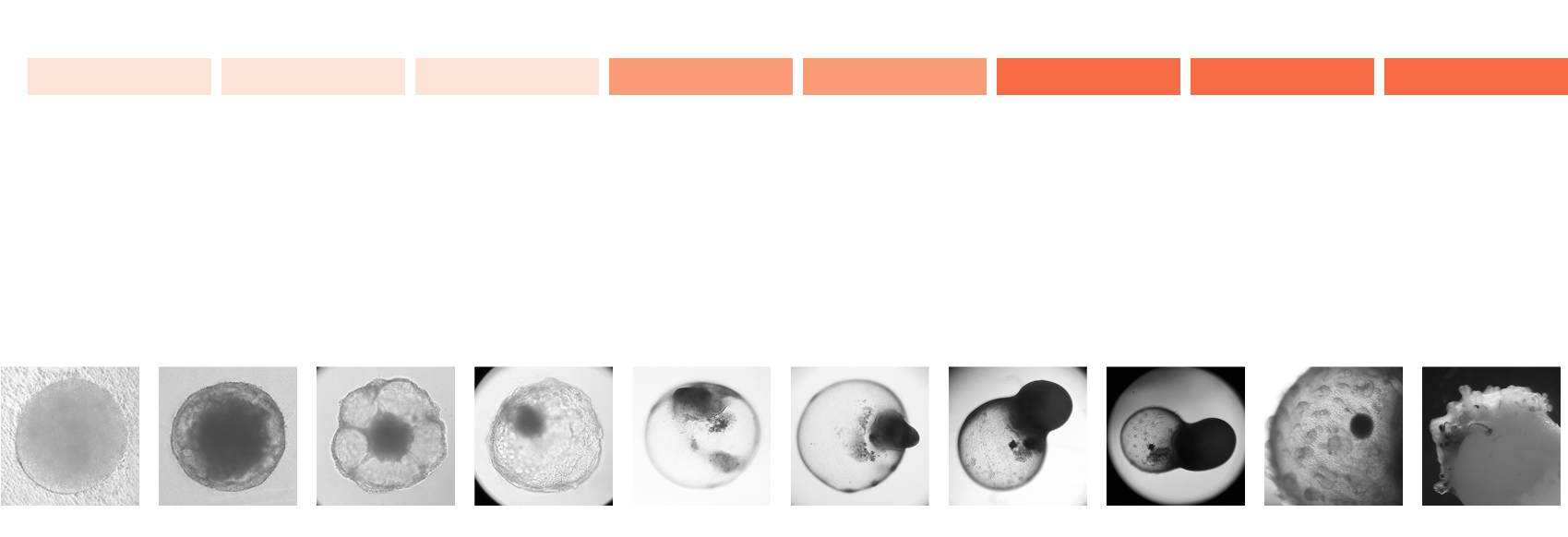
Cellular and structural similarity
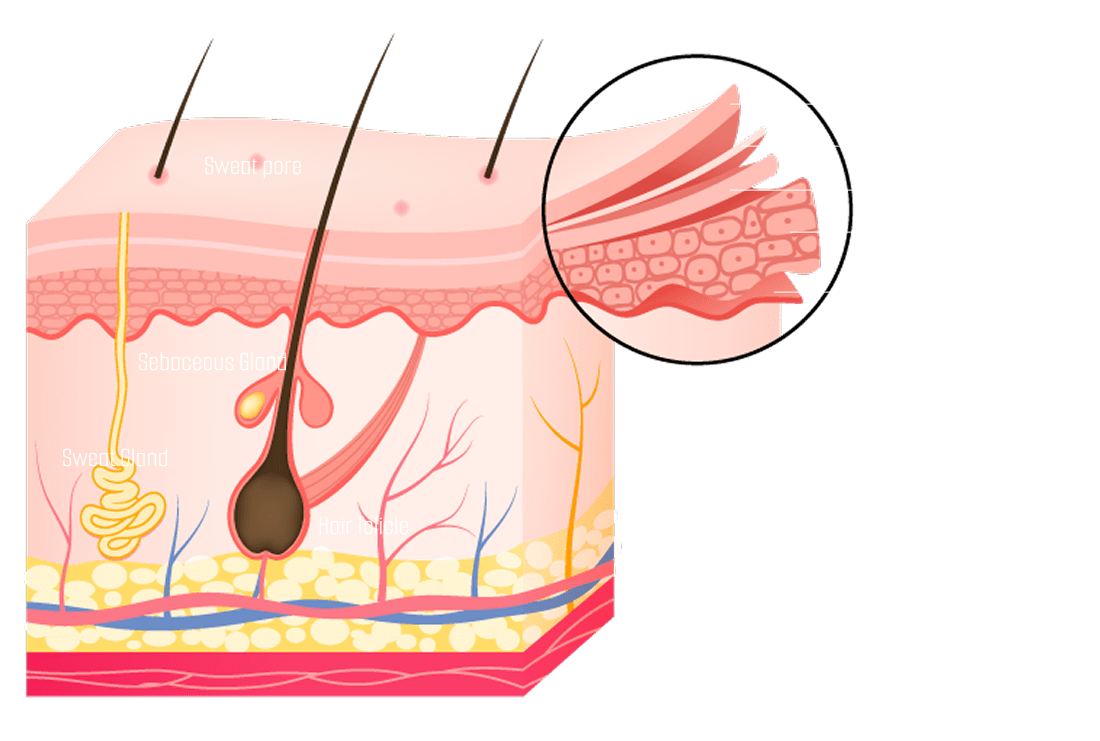
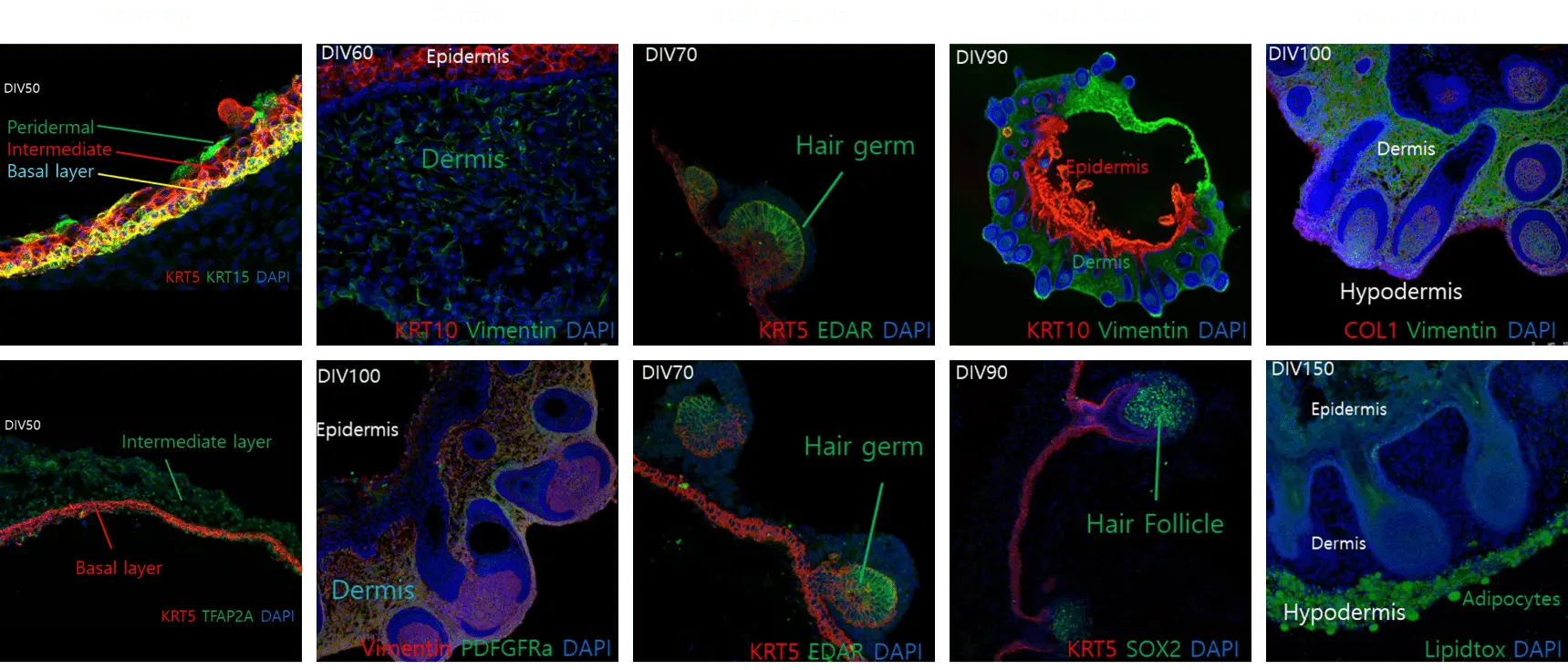
Structural characteristics of skin organoids
Skin Structure
- Expression Markers
Epidermis
- KRT5Basal Layer
- KRT10Spinous Layer
- KRT15Peridermal
- TFAP2AIntermediate Layer
Dermis
- VimentinFibroblast Intermediate Filament
- PDFGFRαMesenchyme- Associated Cell
- COL1Collagen
- EDARHair Germ
- SOX2Hair Follicle
Hypodermis
- LipidtoxAdipocytes
Advanced skin organoid manufacturing technology
Previous method
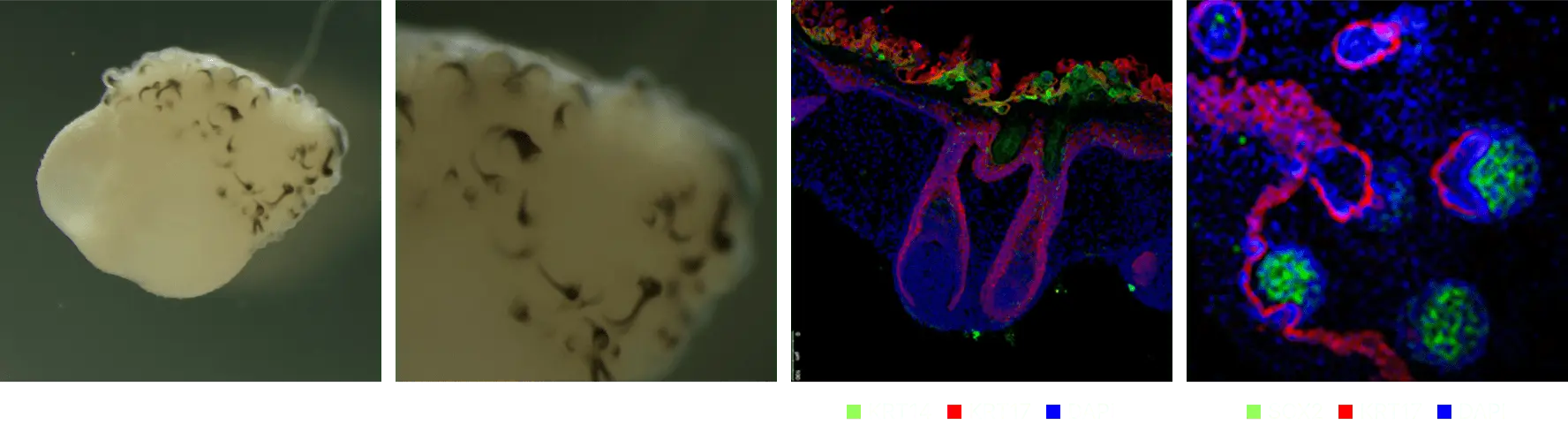
Hypodermis
- KRT5
- KRT17
- EDAR
- SOX2
Skin structure
- Basal Layer [Epidermis]
- Cornified envelope [Epidermis]
- Hair Gem [Dermis]
- Dermal papilla cell [Dermis]
Advanced method
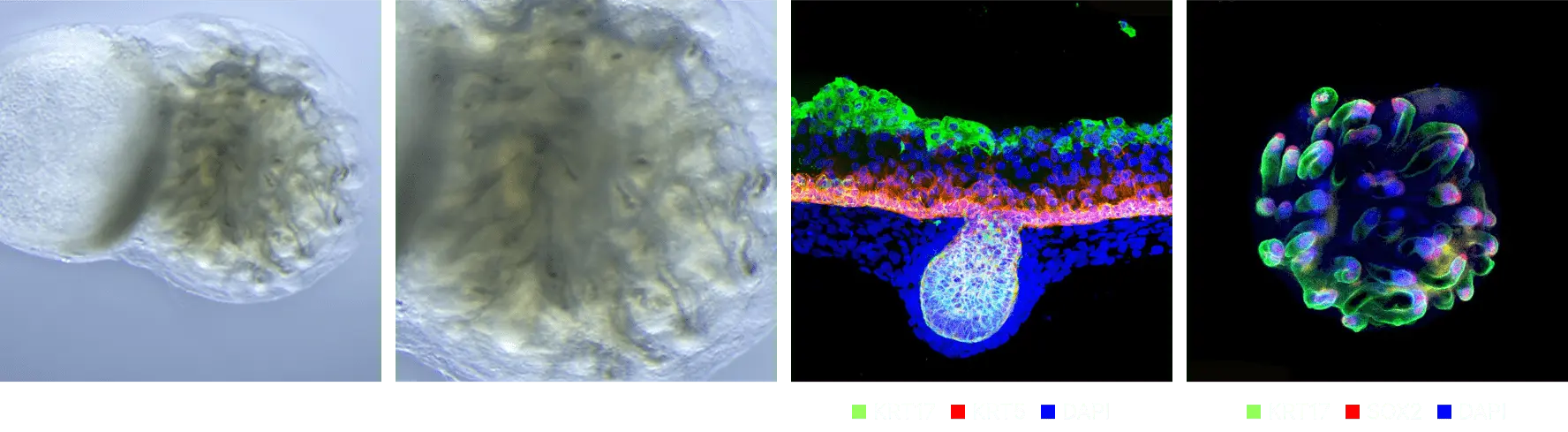
Evaluation of drugs for hair formation effectiveness
Structural characteristics of skin organoids
When treated with an effective drug, an increase in the number of hair follicles was obtained, and additional research on mechanisms related to hair follicle formation was conducted.
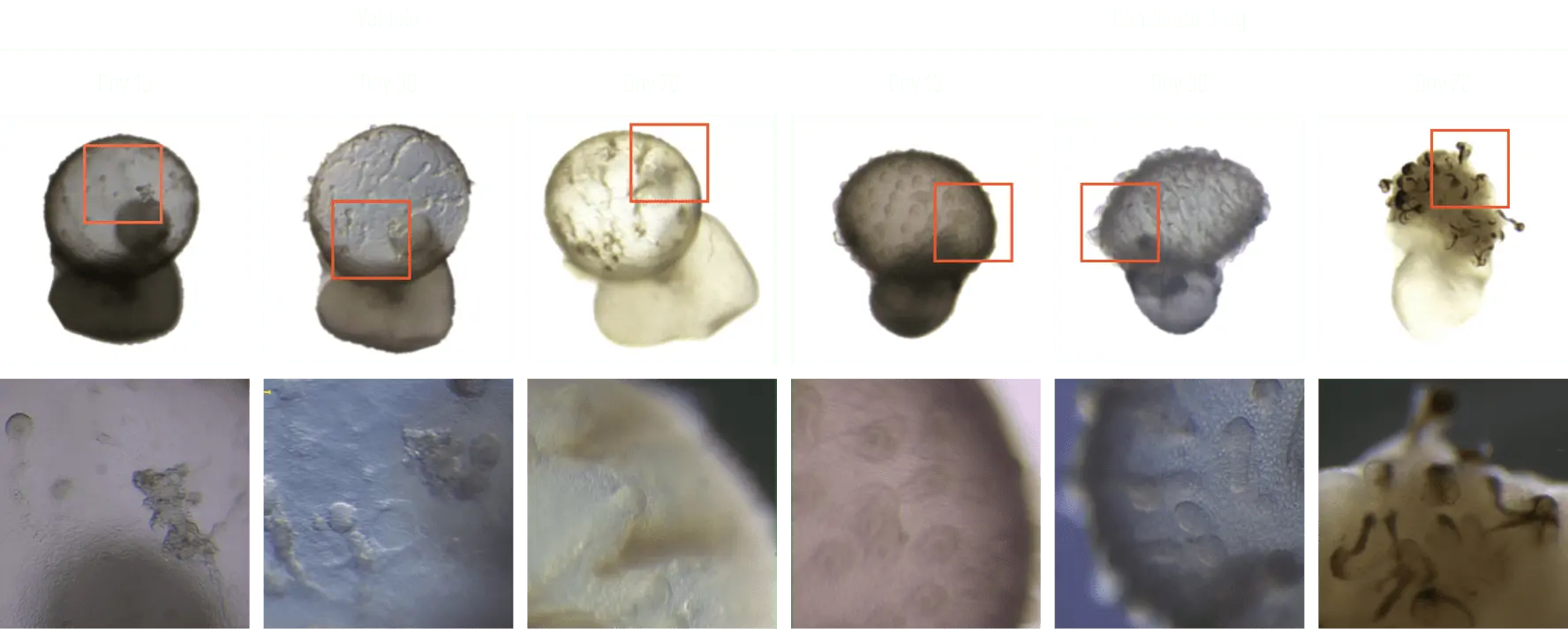
Hair loss model production and effectiveness evaluation
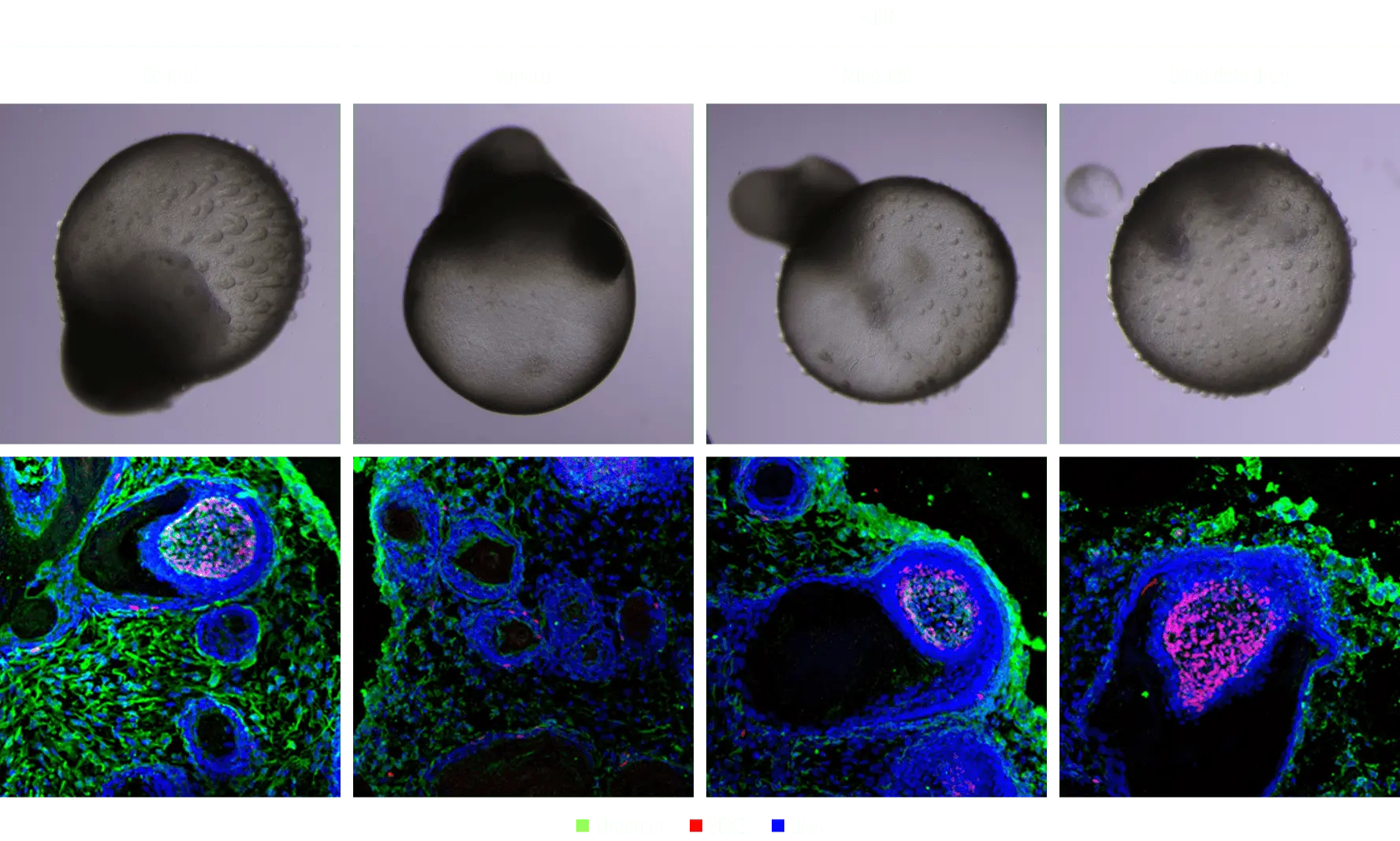
ODISEI Spatial
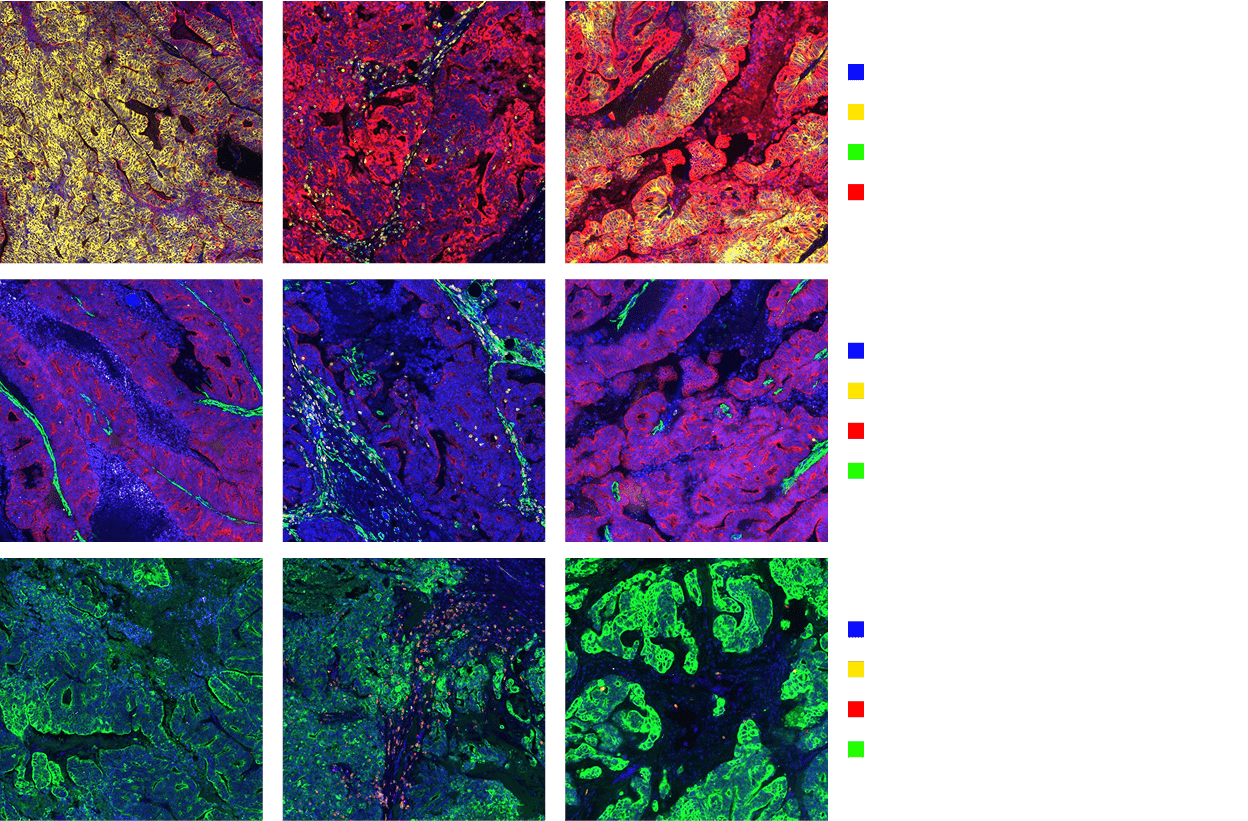
Phenocycler(CODEX)
CODEX was developed to overcome the limitations of existing IHC.
Multiplex-IHC is an experimental method that can confirm the expression of more than 100 biomarkers on one tissue slide.
It is possible to check the expression of various biomarkers on a single tissue slide and conduct comprehensive research on cell composition, cell function and state, and cell-cell interaction.
CODEX analysis is performed using DNA barcoding and uses antibodies conjugated to unique oligonucleotide sequences. By using a target specific barcode called dye-labelled reporter, it is possible to stain more than 100 biomarkers by repeating the labeling cycle by dyeing three colors of fluorescence in one cycle.
Our Core Values
- Staining more than 100 biomarkers on tissue slides 1 chapter.
- Mapping of Millions of Cells Through Diverse Panel Combinations.
- Single cell resolution analysis.
- Cell phenotyping using marker staining combinations.
- Cellular Neighborhood Analysis Utilizing Spatial Information of Cells.
A new standard for Immuno-oncology research has arrived
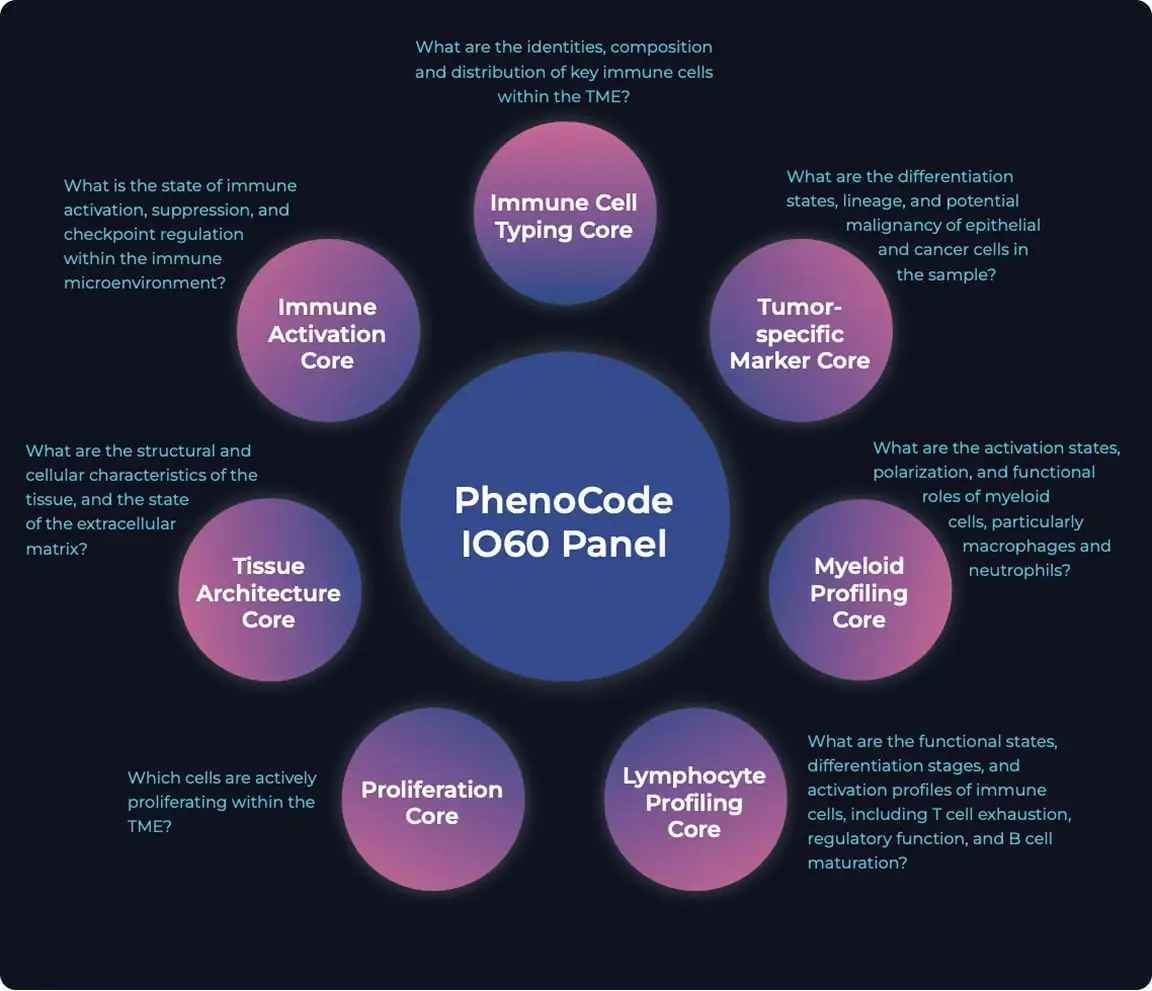
New panel (IO60) Open
Advantage of IO60
- Expanded panel enables deeper profiling of immune-tumor interactions.
- Improved resolution in identifying rare immune subsets and tumor heterogeneity.
- Enhanced capability for co-expression and spatial marker mapping.
- More markers, more insight—accelerate biomarker discovery and therapeutic evaluation.
Immune cell type core
- 1. CD68
- 2. CD4
- 3. CD44
- 4. CD45RO
- 5. CD45
- 6. CD11c
- 7. CD8
- 8. HLA-A
- 9. HLA-DR
- 10. CD14
- 11. CD20
- 12. CD3e
- 13. CD56
Immune activation
- 1. IFNG
- 2. IDO1
- 3. PD-1
- 4. ICOS
- 5. PD-L1
- 6. LAG-3
- 7. VISTA
Tissue architecture
- 1. CD31
- 2. E-cadherin
- 3. SMA
- 4. Vimentin
- 5. CD34
- 6. Beta-actin
- 7. Podoplanin
- 8. Collagen IV
- 9. Caveolin
- 10. B-catenin1
Lymphocyte profiling
- 1. CD107
- 2. CD57
- 3. FOXP3
- 4. CD21
- 5. CD38
- 6. Granzyme B
- 7. TOX
- 8. TCF-1
- 9. CD79a
- 10. CD39
Myeloid profiling
- 1. iNOS
- 2. CD66
- 3. MPO
- 4. CD163
- 5. CD11b
- 6. CD206
- 7. CD209
Proliferation
- 1. Histone H3 Phospho(Ser28)
- 2. PCNA
- 3. Ki67
Tumor-specific marker
- 1. Keratin 14
- 2. SOX2
- 3. Pan-CK
- 4. Keratin 8/18
- 5. ER
- 6. Bcl-2
- 7. EpCAM
- 8. GP100
- 9. TP63
- 10. Keratin 5
The published paper using our platform



Read out
31markers (charged slide)
Teratoma formation analysis
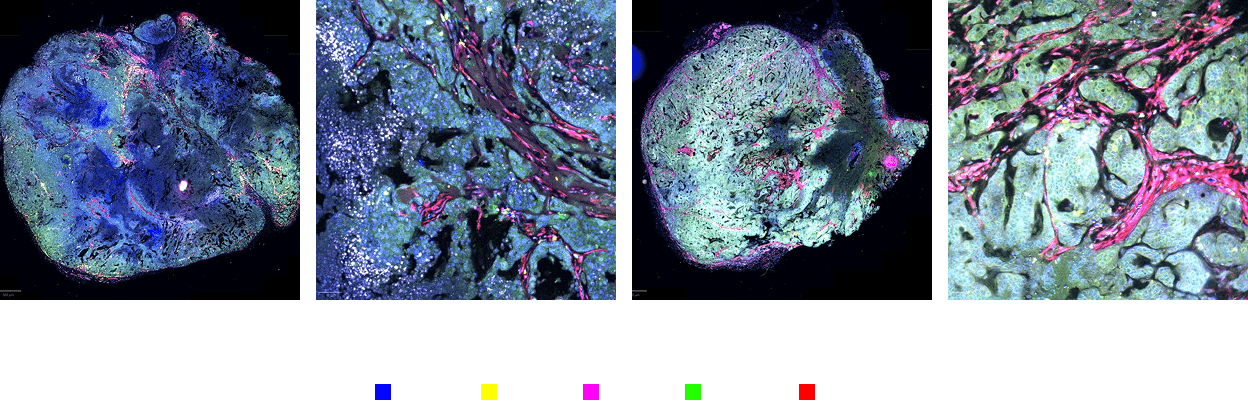
Simultaneous staining of 31 markers in tumor tissue
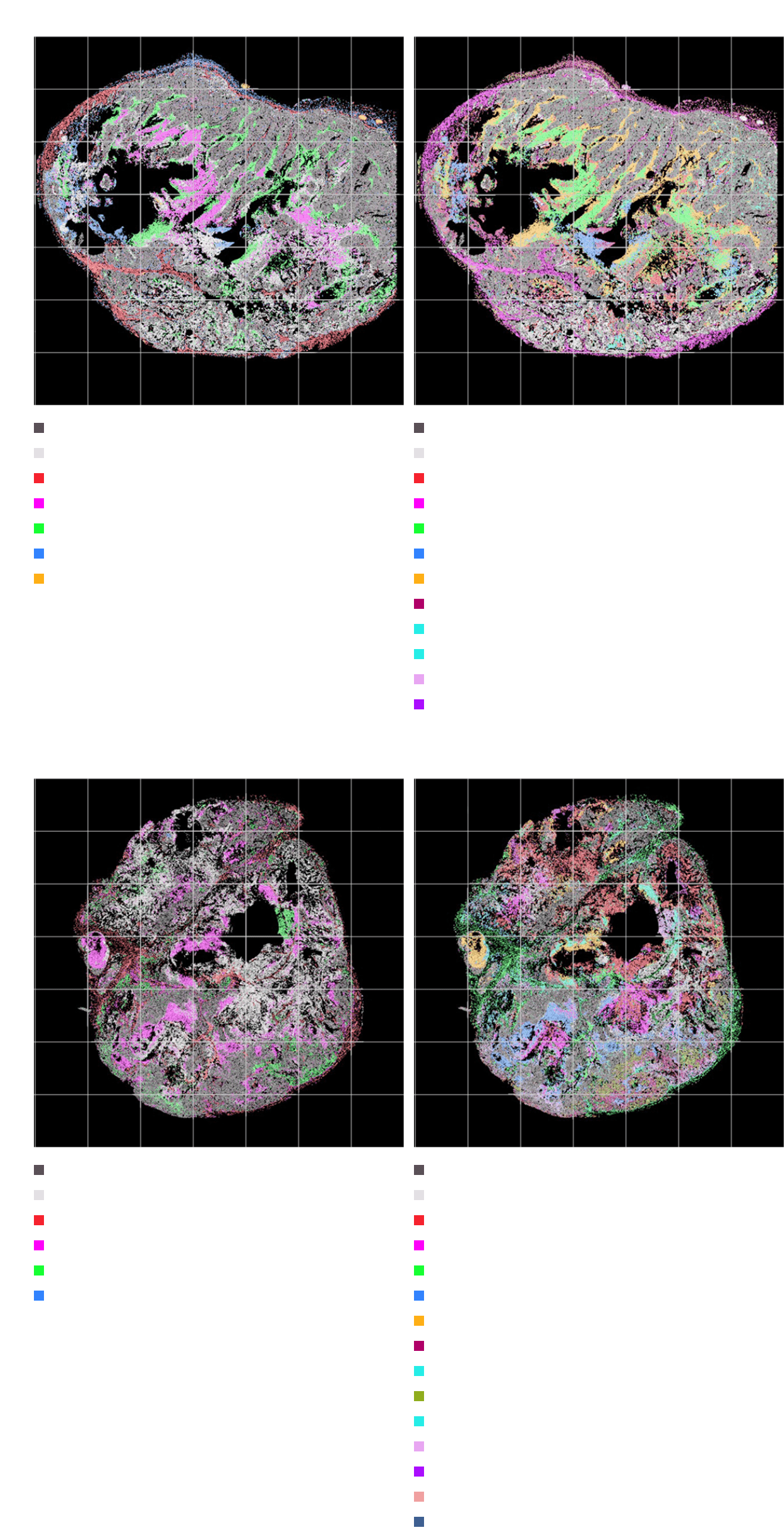
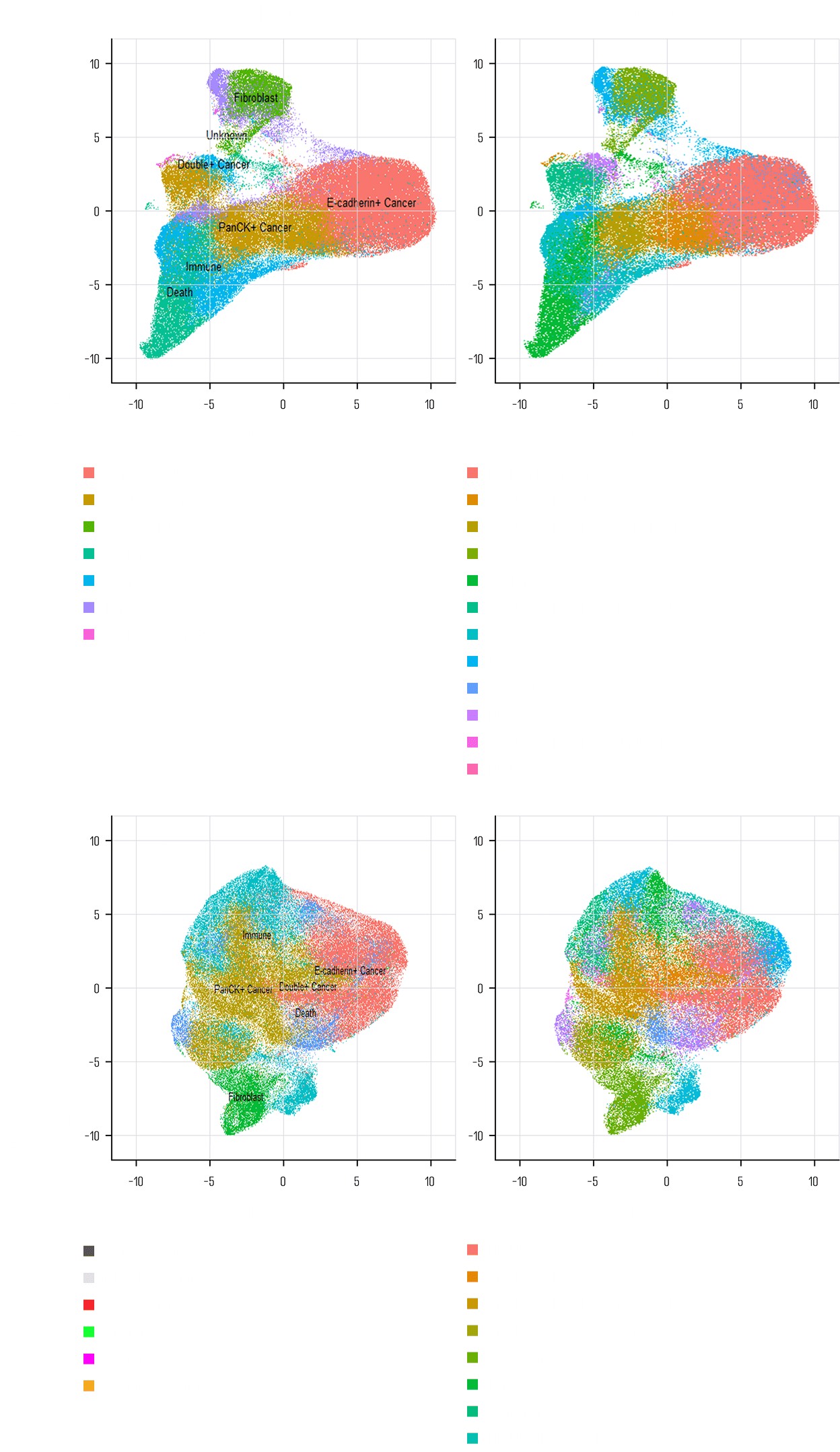
Identification of treatmentmechanism through spatial analysis of cell types
Toward a Sustainable Future with ORGANOIDSCIENCES
Be part of the journey of biomedical innovation that is shaping the future of healthcare.
Technical Inquiry
bd@organoidrx.com
PR/Marketing inquiry
pr@organoidrx.com
Recruitment inquiry
hr.admin@organoidrx.com
General Inquiry
info@organoidrx.com
